Y (male transmitted) Haplogroups according to environment (1 December, 2014, 9 Kislev, 5775)
The Ancient Roman appreciation of the cause of human differentiation is still valid.
Tacitus, from Agricola, pacification of Britain The Works of Tacitus,
11. Who were the original inhabitants of Britain? Whether they were indigenous or foreign is, as usual among barbarians, little known. Some conclusions may be drawn from the inhabitants' various physical traits. The red hair and large limbs of the inhabitants of Caledonia [Scotland] point clearly to a German origin. The dark complexion of the Silures ['People of the Rocks,' southern Wales], their usually curly hair, and the fact that Spain is the opposite shore to them, are an evidence that Iberians [Spanish] of a former date crossed over and occupied these parts. Those who are located nearest to the Gauls are also like Gauls in appearance, either from the permanent influence of original descent, or because, in countries so similarly situated, climate has produced similar physical qualities.
Source: English tr. Alfred John Church and William Jackson Brodribb [1864-1877] at sacred-texts.com, modernized by Dr. G
http://www.sacred-texts.com/cla/tac/ag01010.htm
Note the expression in the last sentence, Native Britons living near the Gauls looked similar to them "either from the permanent influence of original descent, or because, in countries so similarly situated, climate has produced similar physical qualities."
That sums it up.
A similarity in physical type may reflect racial kinship or it may simply show a mutual exposure to the same environmental factors in the past.
What applies to physical type is also applicable to DNA.
Is Not DNA Determined by the Environment???
http://www.britam.org/DNAenvironment.html
DNA studies applied to racial distribution concentrates on mtDNA transmitted exclusively (it used to be thought) by females and on YDNA which is passed on through the male line.
Environmental influence on mtDNA has been admitted for some time.
MtDNA asnd Environment
Thursday, July 09, 2009
Climate shaped the worldwide distribution of human mitochondrial DNA sequence variation
http://yannklimentidis.blogspot.com/2009/07/human-mtdna-subject-to-selection-by.html
Francois Balloux, Lori-Jayne Lawson Handley, Thibaut Jombart, Hua Liu and Andrea Manica
Proceedings Royal Society B
Abstract: There is an ongoing discussion in the literature on whether human mitochondrial DNA (mtDNA) evolves neutrally. There have been previous claims for natural selection on human mtDNA based on an excess of non-synonymous mutations and higher evolutionary persistence of specific mitochondrial mutations in Arctic populations. However, these findings were not supported by the reanalysis of larger datasets. Using a geographical framework, we perform the first direct test of the relative extent to which climate and past demography have shaped the current spatial distribution of mtDNA sequences worldwide. We show that populations living in colder environments have lower mitochondrial diversity and that the genetic differentiation between pairs of populations correlates with difference in temperature. These associations were unique to mtDNA; we could not find a similar pattern in any other genetic marker. We were able to identify two correlated non-synonymous point mutations in the ND3 and ATP6 genes characterized by a clear association with temperature, which appear to be plausible targets of natural selection producing the association with climate. The same mutations have been previously shown to be associated with variation in mitochondrial pH and calcium dynamics. Our results indicate that natural selection mediated by climate has contributed to shape the current distribution of mtDNA sequences in humans.
The study below shows a perceived correspondence between specific YDNA haplogroups and the surroundings.
Hum Biol. 2014 May;86(2):113-30.
Human paternal lineages, languages, and environment in the caucasus.
Tarkhnishvili D1, Gavashelishvili A1, Murtskhvaladze M1, Gabelaia M1, Tevzadze G2.
http://dienekes.blogspot.co.il/2014/11/paternal-lineages-and-languages-in.html
Abstract
http://dienekes.blogspot.co.il/
Publications that describe the composition of the human Y-DNA haplogroup in diffferent ethnic or linguistic groups and geographic regions provide no explicit explanation of the distribution of human paternal lineages in relation to specific ecological conditions. Our research attempts to address this topic for the Caucasus, a geographic region that encompasses a relatively small area but harbors high linguistic, ethnic, and Y-DNA haplogroup diversity. We genotyped 224 men that identified themselves as ethnic Georgian for 23 Y-chromosome short tandem-repeat markers and assigned them to their geographic places of origin. The genotyped data were supplemented with published data on haplogroup composition and location of other ethnic groups of the Caucasus. We used multivariate statistical methods to see if linguistics, climate, and landscape accounted for geographical diffferences in frequencies of the Y-DNA haplogroups G2, R1a, R1b, J1, and J2. The analysis showed significant associations of (1) G2 with wellforested mountains, (2) J2 with warm areas or poorly forested mountains, and (3) J1 with poorly forested mountains. R1b showed no association with environment. Haplogroups J1 and R1a were significantly associated with Daghestanian and Kipchak speakers, respectively, but the other haplogroups showed no such simple associations with languages. Climate and landscape in the context of competition over productive areas among diffferent paternal lineages, arriving in the Caucasus in diffferent times, have played an important role in shaping the present-day spatial distribution of patrilineages in the Caucasus. This spatial pattern had formed before linguistic subdivisions were finally shaped, probably in the Neolithic to Bronze Age. Later historical turmoil had little influence on the patrilineage composition and spatial distribution. Based on our results, the scenario of postglacial expansions of humans and their languages to the Caucasus from the Middle East, western Eurasia, and the East European Plain is plausible.
We have here the following associations:
(1) G2 with wellforested mountains, (2) J2 with warm areas or poorly forested mountains, and (3) J1 with poorly forested mountains.
G2 is part of F.
F is the parent of GHIJKLT
In human genetics, Haplogroup G (M201) is a Y-chromosome haplogroup. It is a branch of Haplogroup F (M89). Haplogroup G has an overall low frequency in most populations but is widely distributed within ethnic groups of the Old World in the Middle East, Europe, Caucasus, South Asia, western and central Asia, and northern Africa.
Its highest concentration is in the Caucasus. followed by the Levant and Iran. Its possible place of origin is the Levant. It has been found in prehistoric western Europe.
It may be linked with relatively mild climates and areas that historically were probably forested.
J2 Italy, Greece, Anatolia, Caucasus, also amongst Jews (ca. 20% but ca. 80% of Cohens). In the Caucasus it was linked with warm areas or poorly forested mountains.
J1 in the Caucasus was linked with poorly forested mountains. J1 is concentrated amongst Arabs who are natives of the desert and sparse vegetation.
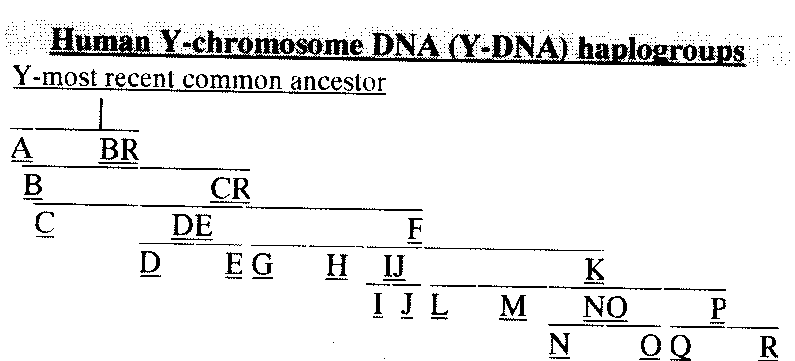
F is the parent of GHIJKLT
G forested areas.
IJ are twins. I is a version of J for colder regions such as the Adriatic coast, Scandinavia, Northern Germany.
J2 is J in warm areas with some vegetation cover and cultivation.
J1 is J in warm areas with little vegetation and sparse cultivation.
L is sparse everywhere. apparent relevant concentration in mountainous areas. Highest (23%) amongst Kalash of northwest Pakistan. Possibly cold relatively dry areas.
T is F in warmer climates.
http://www.eupedia.com/europe/Haplogroup_T_Y-DNA.shtml
Haplogroup T is a fairly rare lineage in Europe. It makes up only 1% of the population on most of the continent, except in Greece, Macedonia and Italy where it exceeds 4%, and in Iberia where it reaches 2.5%, peaking at 10% in Cadiz and over 15% in Ibiza. The maximal worldwide frequency for haplogroup T is observed in East Africa (Eritrea, Ethiopia, Somalia, Kenya, Tanzania) and in the Middle East (especially the South Caucasus, southern Iraq, south-west Iran, Oman and southern Egypt), where it accounts for approximately 5 to 15% of the male lineages. Besides these regions and Europe, T is found in isolated pockets as far as Central Asia, India, Cameroon, Zambia and South Africa. Its highest density is actually found among the Fulani people of Cameroon (18% of the population).
N cold wet stable areas.
Y Haplogroup Q
http://www.eupedia.com/europe/Haplogroup_Q_Y-DNA.shtml
Haplogroup Q is found predominantly in Central Siberia, Central Asia and among Native Americans. Approximately 90% of pre-Columbian Native Americans belonged to haplogroup Q, and all descend from the branch Q1a2a1 (L54), including various subclades of Q1a2a1a1 (M3) and Q1a2a1a2 (Z780). In Europe haplogroup Q is found chiefly in southern Sweden (5%), among Ashkenazi Jews (5%), and is various isolated pockets in central and Eastern Europe such as the Rhone-Alpes region of France, southern Sicily, southern Croatia, northern Serbia, parts of Poland and Ukraine. Sari et al. (2013) also found 6.1% of haplogroup Q out of 412 samples from the island of Hvar in southern Croatia (accompanied by 2% of East Asian mtDNA haplogroup F).
Y haplogroup O appears in 80-90% of all men in East and Southeast Asia, and it is almost exclusive to that region.
R1a temperate continental.
R1b coastline dwellers. wet climates.
http://www.eupedia.com/europe/Haplogroup_J2_Y-DNA.shtml
Y haplogroup E
There are several different clades of Y haplogroup E. This is the major haplogroup of Africa. All clades of E are black-skinned except for E1b1b which is white. E1b1b exists more or less in the intermediate range between white and black areas. Which came first heredity or environment is another question but environment is an undeniable factor.
http://www.eupedia.com/europe/Haplogroup_E1b1b_Y-DNA.shtml
Outside Europe, E1b1b is found at high frequencies in Morocco (over 80%), Somalia (80%), Ethiopia (40% to 80%), Tunisia (70%), Algeria (60%), Egypt (40%), Jordan (25%), Palestine (20%), and Lebanon (17.5%). On the European continent it has the highest concentration in Kosovo (over 45%), Albania and Montenegro (both 27%), Bulgaria (23%), Macedonia and Greece (both 21%), Cyprus (20%), Sicily (20%), South Italy (18.5%), Serbia (18%) and Romania (15%).
The highest genetic diversity of haplogroup E1b1b is observed in Northeast Africa, especially in Ethiopia and Somalia, which also have the monopoly of older and rarer branches like M281, V6 or V92. Ethiopians and Somalians belong mostly to the V22 and V32 (downstream of V12) subclades, but possess also a minority of M81, M123 and V42 subclades. Among the main subclades of E1b1b only V13 and V65 are absent from the Horn of Africa, and probably originated in northern Africa (V65) or the southern Levant (V13).Y haplogroup E
There are several different clades of Y haplogroup E. This is the major haplogroup of Africa. All clades of E are black-skinned except for E1b1b which is white. E1b1b exists more or less in the intermediate range between white and black areas. Which came first heredity or environment is another question but environment is an undeniable factor.
http://www.eupedia.com/europe/Haplogroup_E1b1b_Y-DNA.shtml
Outside Europe, E1b1b is found at high frequencies in Morocco (over 80%), Somalia (80%), Ethiopia (40% to 80%), Tunisia (70%), Algeria (60%), Egypt (40%), Jordan (25%), Palestine (20%), and Lebanon (17.5%). On the European continent it has the highest concentration in Kosovo (over 45%), Albania and Montenegro (both 27%), Bulgaria (23%), Macedonia and Greece (both 21%), Cyprus (20%), Sicily (20%), South Italy (18.5%), Serbia (18%) and Romania (15%).
The highest genetic diversity of haplogroup E1b1b is observed in Northeast Africa, especially in Ethiopia and Somalia, which also have the monopoly of older and rarer branches like M281, V6 or V92. Ethiopians and Somalians belong mostly to the V22 and V32 (downstream of V12) subclades, but possess also a minority of M81, M123 and V42 subclades. Among the main subclades of E1b1b only V13 and V65 are absent from the Horn of Africa, and probably originated in northern Africa (V65) or the southern Levant (V13).
Conclusion:
The Y haplogroups to a greater or lesser degree may on the whole be defined as having a homeland of specific environment.
